Analysis of Integrated Structure of the Exogenous Gene Insertion and the Establishment of Event-specific Detection Method in Transgenic Insect Resistant Cotton 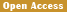
2 Biotechnology Research Institute, CAAS, Beijing, 100081, P.R.China
 Author
Author  Correspondence author
Correspondence author
GMO Biosafety Research, 2012, Vol. 3, No. 1 doi: 10.5376/gmo.2012.03.0001
Received: 04 May, 2012 Accepted: 18 Jun., 2012 Published: 21 Jun., 2012
Hou et al., 2012, Analysis of Integrated Structure of the Exogenous Gene Insertion and the Establishment of Event-specific Detection Method in Transgenic Insect Resistant Cotton, GMO Biosafety Research, Vol.3, No.1 1-7 (doi: 10.5376/gmo.2012.03.0001)
In this study, the complete sequence of exogenous DNA insertion was obtained from transgenic cotton line 06N-119 by using 3'-terminal high-efficiency thermal asymmetric interlaced PCR (hiTAIL-PCR) two times, 5'-terminal hiTAIL-PCR four times and long distance PCR (LD-PCR) once. The insertion of the exogenous gene is 11578 bp in length and consists of the Nptâ…¡ gene expression cassette and the Bt gene expression cassette in series. There is a 75 bp fragment lost at insertion site but no gene recombination occurred. The event-specific qualitative PCR method was established based on the 3'-terminal flanking sequence, which was proved to be highly specific and sensitive. The detection limit was as low as 0.05% every 100 ng of template, that is equivalent 11 target sequence copies. The results of this study would be importance on the detection of exogenous gene in transgenic cotton and biosafety assessment.
Since the transgenic cotton initiated to study in 1989, China has made great achievements in the field of genetically modified cotton research. The domestic transgenic Bt cotton being successful developed in 1994 made China become the second country succeeding in the United States who had independent intellectual property rights of the insect-resistant genes (Cui and Guo, 1996; Mao and Guo, 1998; Huang et al., 1998). With the successful development of the bivalent transgenic cotton harboring Bt and CpTI in 1996 (Guo and Cui, 1998; Cui and Guo, 1998; Guo et al., 1999), which not only delays the bollworm’s resistance to Bt protein but also enhances the toxic to insect, China entered the international advanced level in the field of cotton research. It is said that the Bt cotton made by China occupied for more than 93 percent of China's Bt cotton market in 2010.
In recent years, with the continuous expansion of the transgenic Bt cotton planting area, safety assessment and supervision of genetically modified crops have been maturing in China. Therefore, it has become increasingly urgent to study the integrated structure of the exogenous gene and to establish a scientific testing method in the events of genetically modified crop varieties (lines). Guo and Zhang (2010) conducted a preliminary study to obtain the flanking sequence of left and right borders of T-DNA insertion sites by TAIL-PCR, which the exogenous DNA fragments were transformed by cotton pollen tube pathway approach into; Cui (2004) preliminary analyzed integration flanking sequence of insecticidal gene in the Bt cotton GK12, providing a reference for the integration mechanism of the insecticidal gene; Wang (2011) obtained flanking sequences in EZaMian 1Hao by the Genome Walking combined with the long-chain method, and further screened and identified strain-specific detection primers. However, the complete structure of the inserted fragment did not report yet.
At present, the most popular methods for acquiring the flanking sequence of the foreign gene are the target genome walking PCR (Parker et al., 1991), and thermal asymmetric interlaced PCR ( TAIL-PCR) that developed based on the target genome walking PCR, (Liu and Whitter, 1995) and the high-efficiency TAIL-PCR (hiTAIL-PCR) (Liu and Chen, 2007).
In this study, Using hiTAIL-PCR and long-chain PCR method, we attempted to obtain the flanking sequence of exogenous DNA insertion sites in transgenic cotton 06N-119 in order to elucidate integration structure in detail. While we try to design specific PCR primers based on the flanking sequence of the 3' end of transformants and to conduct qualitative PCR amplification in order to establish the transforming event-specific detection methods.
1 Results and Analysis
1.1 Obtaining 3' flanking sequence of insertion fragment
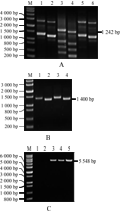
The insertion fragment of the 3' flanking sequence and the cotton genome sequence were obtained by hiTAIL-PCR amplification experiments twice, and then PCR validation was done. First, using 3-1sp1, 3-1sp2 and 3-1sp3 as specific primers, a 2776-bp fragment was obtained by hiTAIL-PCR amplification experiments using the sample DNA as template, (Figure 1A), and then a long-range PCR validation was carried out (primers 5'-F1/3'-R4) (Figure 1B). Bioinformatics analysis showed that the 2200 bp of the fragment was the cry1Ac gene sequence, while the 200 bp was the 7S-UTR, which were similar with the sequences of Xinmian 33B.
The three specific primers, named 3-2sp1, 3-2sp2, and 3-2sp3 were designed based on the obtained fragments of 2776 bp to conduct the second hiTAIL-PCR amplification, a 1118 bp fragment was obtained. BLASTn analysis showed that 200 bp of the fragment was 7S-UTR sequence, and 800 bp of the fragment was no any match of homologous sequences. The forward primer, named MF-1 was designed according to the 7S-UTR sequences, while reverse primer named MR-5 was designed based on 800-bp sequence. PCR validation was conducted to generate 926 bp fragment of the sample 06N-119, while there is no amplified band appearing in the positive and negative controls, (Figure 1C). The primers (MF-3/MR-5,) were designed based on the region of 800 bp sequence, the PCR result showed that both of the cotton samples and controls can be amplified the 506 bp band, which indicated that the obtained 800 bp fragment are the sequence of the cotton genomic sequence (Figure 1D).
|
Figure 1 The insertion fragment of GM cotton 06N-119 amplified by 3′-terminal hiTAIL-PCR
|
1.2 Obtaining the 5' end flanking sequence of insert fragment
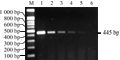
The 5' flanking sequence of the insert fragment and the cotton genome sequence were obtained by hiTAIL-PCR and a long distance PCR amplification experiments four times. The hiTAIL-PCR amplification was performed four times toward the 5' end sequence of the cry1Ac gene. A 1242 bp fragment was amplified at the first amplification (Figure 2A), which contained 200 bp of cry1Ac sequence, 521 bp of the 35S promoter sequence, 500 bp of the vector backbone; A 1678 bp fragment was amplified at second amplification, which contained 276 bp of NOS terminator, 600 bp of the Npt II gene sequence, and the remaining of the vector backbone; A 690 bp fragment was obtained at the third amplification, which contained 195bp of Npt II gene sequences and 400 bp of the E35S promoter sequences.
Because the follow-up hiTAIL-PCR amplification experiments failed to get the specific bands, we took the following method to acquire the further 5' flanking sequence. The direction of hiTAIL-PCR amplification toward the direction of the 5' end sequence was carried out by sing the negative non-genetically modified cotton Ji 668 genomic DNA as a template, and specific primers named c-1sp1, c-1sp2 and c-1sp3 designed based on the obtained 3' end cotton genomic sequence, a 1400 bp cotton genome sequence was obtained (Figure 2B), then the forward primer (5'-F5) near the 1400 bp sequence and the reverse primer (3'-R3) in the Npt II gene sequences were designed to generate a 5 548 bp band (Figure 2C). The result of sequencing and splicing showed that we obtained the remaining unknown sequence of the 5' end of the insert and 894 bp of cotton genome sequence.
|
Figure 2 The insertion fragment of GM cotton 06N-119 amplified by 5′-terminal hiTAIL-PCR
|
1.3 The structural characteristics of the insert
By conducting the 3' and 5' end hiTAIL-PCR amplification and long distance PCR amplification, we obtained the entire sequence of the insert and the cotton genome sequence of both flanking sides. Bioinformatics analysis showed that exogenous inserting DNA was 11578 bp in full length consisting of two tandem expression cassettes, namely Npt II gene expression cassette and cry1Ac expression cassette (Figure 3). The structure of the inserting fragment from 5' to 3' end followed by: (1) ori322/629 bp; (2) oriV/394 bp; (3) E35S promoter/521 bp; (4) Npt II/795 bp ; (5) NOS terminator/254 bp; (6) coding region/789 bp; (7) E35S promoter/521 bp; (8) cry1Ac/3537 bp; (9) 7S-UTR/416 bp. The results further indicated that the promoter for Npt II gene was the enhanced promoter E35S, which was fully consistent with the promoter sequences of the cry1Ac gene. The cry1Ac gene cassette structure between cotton 06N-119 and Xinmian 33B was complete identical, but 8 base of the cry1Ac gene in 06N-119 was different with the XInmian 33B. The specific primer for Mon531 line (Yang et al., 2005) was employed to detect on the 06N-119, the results failed to detect the specific bands, indicating that Mon531 line-specific detection method should not be suitable for the detection of 06N-119.
Comparison between the cotton genomic sequences of the sides of the insert fragments in 06N-119 and the same regional of cotton genomic sequences in non-transgenic cotton Ji 668, we found that a 75 bp cotton genomic fragment was lost during the insertion event occurred, but did not found gene recombinant phenomena in the insertion site region (Figure 3).
|
|
1.4 Establishment of specific PCR detection method for 3' end of the transformation event
In order to establish the specific PCR detection method for 3' end of the transformation event, we used genomic DNA as template to screen the primes and their combinations based on the amplification efficiency and specificity, total of 10 combinations among the five forward primers and six reverse primers were screened to determine the MF-1/MR-2 as the 3' end specific detection primers of genetically modified cotton. Then using this screened primers to detect the specificity on different crops, the results showed that only 06N-119 contained the expected 445 bp band, while the other 32 samples including cotton, rice, corn, soybean, rapeseed, sugar beet, and wheat were unable to amplify target band, which indicated that the establishment of transformant specific qualitative PCR detection method for cotton 06N-119 should be high specificity.
The genomic DNA of GM 06N-119 were gradient diluted by using the non-transgenic gene cotton genomic DNA (100 ng of/μL), the diluted concentration were respectively 10.00%, 1.00%, 0.50%, 0.10%, 0.05% and 0%, which used as templates for PCR amplification with MF-1/MR-2 primer pair. The sensitivity testing results showed that the primer pair MF-1/MR-2 could still amplify the specific target fragment in genomic DNA concentration of 06N-119 that was diluted to 0.05% (Figure 4), which indicated that the MF-1/MR-2 detection sensitivity should be at least 0.05% level, namely about 11 copies.
|
Figure 4 Sensitivity detection for event-specific primer pair MF-1/MR-2
|
2 Discussions
Wang et al (2011) found that the transgenic cotton EZaMian 1 Hao share the same construct with Monsanto's Xin Mian 33B, but the transformation event is not the same as the Monsanto’s. By detecting the 5' end flanking sequence of EZAmian 1 Hao, the 5' end of strain-specific detection method was established, but it has not yet to report the 3' end flanking sequence. In this study, we obtained the complete structure of the insertion of transgenic cotton 06N-119 and 5' end and 3' flanking sequence, in which the 5' flanking sequence of EZAmian 1 Hao was complete identical to that reported by Wang et al (2011).
At present, the cotton genome sequencing is in progress. In this study, although insert fragment flanking sequence of 5' end 894 bp and 3' end 800 bp in transgenic cotton 06N-119 did not yet search for homologous sequences in GenBank, it was proved to be the cotton genome sequence by blasting the region of the negative control sequence. These insertion sites might map on the chromosome once the cotton whole genome sequencing is completed.
According to the study of Pan et al (2003), the sensitivity of the primer detection reaching 0.1% can meet the requirements for detecting genetically modified crop. On the basis of the 3' end flanking sequence, 3' end of the transformation event-specific detection methods of the transgenic cotton 06N-119 was established. PCR sensitivity analysis showed that the sensitivity of primers MF-1/MR-2 was 0.05%, which the detection limit was equivalent to 11 exogenous DNA insertion copies. Therefore, the 3' end of the qualitative PCR primer screened in this study had very good stability, strong specificity, and high sensitivity, which would be suitable for specific detection of genetically modified cotton varieties (lines), and might improve and enrich the methods for transgenic Bt cotton transformants specific detection as well as provide a reference for safety testing of transgene.
3 Materials and Methods
3.1 The experimental materials and enzymes and reagents
Transgenic cotton material 06N-119 and other tested varieties such as Zhongmiansuo 41, GK12, and non-genetically modified cotton Ji 668 are cultivated or possessed in this laboratory.
ExTaq DNA polymerase, LATaq DNA polymerase were purchased from TaKaRa Company; GoTaq DNA polymerase purchased from Promega; DNA molecular weight marker, dNTP were purchased Beijing Quanshijin Biotechnology Co., Ltd.; primers were synthesized by Sangon (Shanghai) limited company; other biochemical reagents are imported from abroad for repackaging or domestic analytical grade.
3.2 DNA extraction method
The genomic DNA of young cotton leaves was extracted by using the CTAB method. DNA purity and concentration was determined by UV spectrophotometer, DNA solution was diluted to 50 ng/μL and 100 ng/μL ready for use.
3.3 Obtaining the flanking sequence of the trans- genic cotton
The 5' and 3' end of the flanking sequence of transgenic cotton were determined following the method hiTAIL-PCR of Liu et al (2007) combined with long distance PCR. PCR products were sequenced by Beijing Saino Genome Research Center Co., Ltd., and the sequence alignment were completed with CExpress software and then blasted in BLASTn.
3.4 HiTAIL-PCR reaction and primer designing
HiTAIL-PCR reaction system, procedure and random primer designing followed the literature (Liu and Chen, 2007). 5' and 3' end specific primers were designed based on Cry1Ac sequence; the 5' end specific primers were designed based on the cotton genome sequence. The specific primers of 5' and 3' flanking sequence for amplification were shown in Table 1.
|
Table 1 Specific primer sequences for the hiTAIL-PCR
|
3.5 Long distance PCR (LD-PCR)
To verify the specificity of hiTAIL-PCR products, forward and reverse primers were designed according to the spliced sequence. Long-distance PCR amplification was carried out by using different primer combinations. The reaction system was as follows, F/R primers 1.0 μL (primer concentration 10 μM) LATaq of DNA polymerase 0.25 μL (5 U/μL), 10× LA PCR buffer 2.5 μL, dNTPs 4.0 μL (2.5 mM), 50 ng/μL cotton genomic DNA 1 μL and double distilled water to make up the volume to 25 μL. Reaction procedures was as follows, Pre-denaturation at 94℃ for 1 min in advance, and then 30 amplifying cycles with 98 ℃ 10 s and 66 ℃ 15 min; finally extension at 72 ℃ for 10 min. 5' and 3' end of primer sequences of cry1Ac gene for LD-PCR are shown in Table 2.
|
Table 2 The primer sequences for LD-PCR
|
3.6 The specific PCR detection method for 3' end of the transformation event
3.6.1 The specific verification
The specificity verification of PCR amplification was carried out by employing the genomic DNAs extracted from 12 GM rice samples, 4 GE cotton samples, 4 GE soybean samples, 4 GE oilseed rapes, 2 GE wheat samples and 1 GE beet PCR products were examined by 2% agarose gel electrophoresis.
The specific PCR primers based on between exogenous insertion sequences close to the 3' flanking designed to detect the transformation events. The primers MF-1 (5'-TGTGTACTTCAACTGTCTGCTTAGC-3') and MR-2 (5'-CCCATCTTCTATCCAATCTAACCTC-3') were screened to detect the specificity, while the primers MF-3 (5'-GAGGTTAGATTGGATAGAAGATGGG-3') and MR-5 (5'-ATTCAAGAACTCCTTGGAGGTTGT-3') were screened to verify the cotton genome. PCR Reaction system was set as: 5 μL 5 × GoTaq buffer, 2 μL dNTP (2.5 mmol/L) each primer 0.5 μL (10 μmol/L), 0.13 μL GoTaq (5 U/μL), 1μL Templates (50 ng/μL), adding ultra-purified water up to total 25 μL. PCR amplification procedures were as follows, Pre-denaturation at 94℃ for 5min then 35 amplifying cycles with 94℃ for 30s, 60 ℃ for 30 s, 72 ℃ for 30 s), finally extension at 72 ℃ for 7 min.
3.6.2 Sensitivity test
The GM sample DNA solutions with the concentrations in weight percentage as 10.00% 1.00% 0.50% 0.10% 0.05% and 0% were prepared by using 100 ng/μL genetically modified cotton samples DNA and 100 ng/μL non-transgenic cotton DNA to conduct PCR amplification with the transformation event-specific primers; the sensitivity of the test-specific primers were determined by 2.0% agarose gel electrophoresis. PCR reaction system was as follows, 5 μL 5 × GoTaq buffer, 2 μL dNTP (2.5 mmol/L), each primer 0.5 μL (10 μmol/L), 0.13 μL GoTaq (5U/μL), 1 μL Templates (100 ng/μL), adding ultra-purified water up to total 25 μL. PCR amplification procedures were as follows: Pre-denaturation at 94 ℃ for 5 min then 35 amplifying cycles with 94 ℃ for 30 s, 60 ℃ for 30 s, 72 ℃ for 30 s), finally extension at 72 ℃ for 7 min.
Authors' Contributions
NH carried out the experiments, analyzed the data and wrote the manuscript; MD, RX, and YW conducted a few data analyses and took an active part in experimental design method; JWJ and HBL conceived the experimental design and modified the manuscript. All authors have read and approved the final manuscript.
Acknowledgments
All the authors appreciated two anonymous reviewers for their useful critical comments and revising advice to this paper.
References
Cui H.Z., 2004, The expression of vgbM gene in plants and the preliminary analysis of the cry1Ac gene integration in GK12 cotton, Dissertation for Ph.D., Chinese Academy of Agriculture Science, Supervisor: Guo S.D., pp.75-87
Cui H.Z., and Guo S.D., 1996, Important progress on study of insect-resistance of transgenic cotton in China, Zhongguo Nongye Kexue (Scientia Agricultura Sinica), 29(1): 93
Cui H.Z., and Guo S.D., 1998, Construction of plant expression vectors harboring two insecticidal genes and their expression in tobacco, Nongye Shengwu Jishu Xuebao (Journal of Agricultural Biotechnology), 6(1): 7-13
Guo J.Y., and Zhang T.Z., 2010, Study on the mechanism of pollen-tube pathway-mediated transformation method with TAIL-PCR, Hebei Nongye Daxue Xuebao (Journal of Agricultural University of Hebei), 33(4): 5-9
Guo S.D., Cui H.Z., Xai L.Q., Wu D.L., Ni W.C., Zhang Z.L., Zhang B.B., and Xu Y.J., 1998, New progress on insect-resistant transgenic cotton research in China, Zhongguo Nongye Kexue (Scientia Agricultura Sinica), 31(6): 1
Guo S.D., Cui H.Z., and Xia L.Q., 1999, Development of bivalent insect-resistant transgenic cotton plants, Zhongguo Nongye Kexue (Scientia Agricultura Sinica), 32 (3): 1-7
Huang Q.M., Mao L.Q., Huang W.H., and Guo S.D., 1998, Transgenic tobacco plants with a fully synthesized GFM Cry1A gene provide effective tobacco bollworm (Heliothis Armigera) control, Zhiwu Xuebao (Acta Batanica Sinica), 40(3): 228-233
Liu Y.G., and Chen Y.L., 2007, High-efficiency thermal asymmetric interlaced PCR for amplification of unknown flanking sequences, Biotechniques, 43(5): 649-650, 652, 654, 656
http://dx.doi.org/10.2144/000112601 PMid:18072594
Liu Y.G., and Whitter R.F., 1995, Thermal asymmetric interlaced PCR: automatable amplification and sequencing of insert end fragments from P1 and YAC clones for chromosome walking, Genomics, 25(3): 674-681
http://dx.doi.org/10.1016/0888-7543(95)80010-J
Mao L.Q., and Guo S.D., 1998, Relationship between Ω As well as the length of 3′poly (dA) and efficiency of gene expression, Zhiwu Xuebao (Acta Batanica Sinica), 40 (12): 1166-1168
Pan A.H., Zhang D.B., Pan L.W., Chen J.H., Yuan Z., and Liang W.Q., 2003, A practical PCR method for detection of glyphosate resistant transgenic rape, Zhongguo Nongye Kexue (Scientia Agricultura Sinica), 36 (7): 856-860
Parker J.D., Rebinovitch P.S., and Burmer G.C., 1991, Targeted gene walking polymerase chain reaction, Nucleic Acids Res., 19(11): 3055-3060
http://dx.doi.org/10.1093/nar/19.11.3055 PMid:2057362 PMCid:328270
Wang Q., Xie J.J., Su C.Q., Sun Y., and Peng Y.F., 2011, The Genome Flanking Sequence and Event-specific Qualitative Detection of the transgenic cry1Ac cotton ezamian 1, Nongye Shengwu Jishu Xuebao (Journal of Agricultural Biotechnology), 19(3): 427-433
Yang L.T., Pan A.H., Zhang K.W., Yin C.S., Qian B.J., Chen J.X., Huang C., and Zhang D.B., 2005, Qualitative and quantitative PCR methods for event-specific detection of genetically modified cotton Mon 1445 and Mon531, Transgenic Res., 14(6): 817-831
http://dx.doi.org/10.1007/s11248-005-0010-z PMid:16315089
. PDF(322KB)
. FPDF
. HTML
. Online fPDF
Associated material
. Readers' comments
Other articles by authors
. Na Hou
. Huiqun He
. Mei Dong
. Rongqi Xu
. Yusong Wan
. Wujun Jin
. Haobao Liu
Related articles
. Transgenic resistant cotton
. Integrated structure
. PCR detection
. Event-specificity
Tools
. Email to a friend
. Post a comment