2 Research Institute of South Sichuan, Sichuan Academy of Forestry, Chengdu, 311400, China
3 Ecological Safety on the Upper Reaches of the Yangtze River, National Forestry and Grassland Administration Key Laboratory of Forest Resources Conservation, Sichuan Provincial Key Laboratory of Ecological Forestry Engineering on the Upper Reaches of the Yangtze River, Institute of Ecology & Forestry, Sichuan Agricultural University, Chengdu, 611130, China
 Author
Author  Correspondence author
Correspondence author
International Journal of Molecular Evolution and Biodiversity, 2022, Vol. 12, No. 3 doi: 10.5376/ijmeb.2022.12.0003
Received: 20 Apr., 2022 Accepted: 28 Apr., 2022 Published: 05 May, 2022
Mo J.Y., Peng J., Gu Y.J., and Li X.Q., 2022, Study on population genetic diversity among Picea brachytyla in Jiuzhaigou County, International Journal of Molecular Evolution and Biodiversity, 12(3): 1-11 (doi: 10.5376/ijmeb.2022.12.0003)
To study the genetic diversity and sampling strategy of natural Picea brachytyla populations and provide references for protecting their genetic diversity and resource utilization. In this study, the genetic diversity of six natural P. brachytyla population (90 individuals) were analyzed by genotyping-by-sequencing (GBS). The results showed that six populations could divided into three subsets with high, medium, and low elevation. POP1 belonged to low elevation subset, and mainly obtained individuals in TPG and JWC. POP2 belonged to medium elevation subset, which mainly obtained individuals in QZG and YLG. POP3 belonged to high elevation subset, and mainly obtained individuals in SXC and GGL. SXC was between subset POP2 and POP3, which had genetic communication with the two subsets. There were low genetic differentiation coefficient among six populations, showed a low level of genetic differentiation. The PIC value were all higher than 0.900, indicated that there were highly polymorphism in six populations. The observe heterozygosity and expect heterozygosity were 0.232~0.244 and 0.190~0.199, respectively, and there were no difference between populations, indicated that there remained low level of genetic diversity in P. brachytyla natural populations. The results of correlation analysis showed that there were negative correlation between genetic diversity and altitude, and latitude, and there were no correlation between genetic diversity and longitude. The genetic diversity were decreased with the increase of altitude (from JWC to GGL). This study would provide basis for P. brachytyla genetic conservation. The collection of resources in low altitude would be necessary in germplasm conservation of P. brachytyla.
Picea brachytyla belongs to Picea genus in the family of Pinaceae. It is an endemic tree species in China and is a National Class III Key Protected Plant (Fu, 1992, Science Press, pp. 388-389). Picea brachytyla is an evergreen tree with a height of 40 m, straight trunk and excellent material. It is not only a special aviation material, but also a raw material for construction, furniture and high-grade paper (Fu, 1992, Science Press, pp. 388-389; Botanical Society of China, 1996, China Agricultural Science and Technology Press, pp. 30). More importantly, Picea brachytyla is mainly distributed in alpine areas above 2000 m above sea level. It plays an inestimable role in water conservation and soil and water conservation. It is an excellent tree species for forest regeneration and afforestation in alpine areas. The original distribution center of Picea brachytyla is the mountainous area of Minjiang River and Qingyi River Basin in Sichuan. It is mainly distributed in Pingwu and Songpan in Sichuan. It also distributed in Baoxing and Tianquan along Minjiang River Basin in the south, Hongyuan in the northwest and Kangding in the southwest (Yang, 1992, China Forestry Publishing House, pp. 376-387; Zhou, 1997, Journal of Sichuan Forestry Science and Technology, 18 (4): 19-23). Picea brachytyla is vertically distributed at an altitude of 1800~3000 m. It is a tree species with low altitude distribution of Picea genus in Sichuan Province (Zhou et al., 1997, Journal of Sichuan Forestry Science and Technology, 18 (4): 19-23). Picea brachytyla is a precious tree species left over from the Tertiary period. For a long time, driven by economic benefits, affected by diseases and pests, climate and environment, and lower altitude distribution than that of other Picea plants, the natural forest of Picea brachytyla has been cut down in large quantities, the resources have been reduced sharply, and the loss of germplasm resources has been serious. In addition, there are few studies on population genetic diversity and forest breeding of Picea brachytyla, which makes the research on forest breeding, product research and development and application lag, and seriously affects the development and utilization of Picea brachytyla. Therefore, in this study, Jiuzhaigou area with concentrated natural population distribution and rich species diversity was selected to explore the genetic diversity of Picea brachytyla.
The genetic diversity level of plant population can be accurately evaluated by molecular marker technology (Lu et al., 2018, Lyu and Guo, 2018). As an accurate and efficient new molecular marker technology, genotyping-by-sequencing (GBS) is widely used in the research of plant genetic diversity and other fields (Poland et al, 2012). Genotyping-by-sequencing (GBS) is to interrupt genomic DNA by restriction endonuclease and conduct high-throughput sequencing of specific fragments to obtain a large number of genetic diversity tag sequences, and obtain single nucleotide polymorphisms (SNP) through sequence alignment between individuals or populations, and analyze the genetic diversity and genetic evolution mechanism of species at the genomic level (Yuan, 2011, Science Press, pp. 87-93). For example, Liu (2019) used GBS technology to perform genotyping-by-sequencing on 15 Sophora moorcroftiana populations from different geographical locations. It was found that the genetic diversity of Sophora moorcroftiana in the Yalu Tsangpo River Basin was positively correlated with altitude and negatively correlated with latitude. Wang et al. (2019) used GBS technology to study the genetic diversity of natural population of Litsea populifolia Hemsl. at different altitudes in Emeishan area. It was found that the genetic diversity of natural population of Litsea populifolia Hemsl. in Emeishan area was at a low level, but it had high nucleotide diversity within the population. Taranto et al. (2016) screened 105 184 high-quality SNP loci and divided 222 cultivated varieties of Capsicum annuum L. into two groups.
Jiuzhaigou, located in the southern part of Minshan Mountain, is an important area with dense distribution of Picea brachytyla. Several provincial and national nature reserves were established earlier in Jiuzhaigou, which makes the preservation of Picea brachytyla germplasm resources more perfect and rich. Therefore, Jiuzhaigou is a representative area of Picea brachytyla natural distribution, which is suitable for carrying out research on genetic conservation of Picea brachytyla. Therefore, this study takes the natural population with concentrated distribution of Picea brachytyla in Jiuzhaigou County as the research material, and uses the simplified genome sequencing technology to study its genetic diversity, in order to provide a basis for the genetic conservation of Picea brachytyla. At the same time, it is of great significance for the rational development and utilization of Picea brachytyla and the research of germplasm resources.
1 Results and Analysis
1.1 DNA sample preparation, GBS library construction and SNP detection
Agarose gel electrophoresis of the genomic DNA of Picea brachytyla (Figure 1) showed that the extracted genomic DNA of Picea brachytyla was clear, the quality was high, and the average concentration of Qubit was 121 ng/μL, which accorded with the density requirement of building library. The amount of data sequenced for each sample of 90 Picea brachytyla samples was 1.2~2.6 Gb. After removing the low-quality sequences, the sequenced sequence quality was Q30≥90.26%, the sequencing quality was high, the average GC content was 38.59, and the GC distribution was normal, indicating that the database construction and sequencing was successful. The YLG09 samples with the most tags (19 592 776 in total) were selected for stack clustering and the quasi reference sequences were constructed. Through comparison and detection with SAMTOOLS software, the average comparison rate between the samples to be tested and the quasi reference sequences was 87.39%, indicating that the similarity between each sample and the quasi reference genome met the requirements of re sequencing analysis. A total of 373 291 SNP sites were obtained through comparison with the quasi reference sequences, and 26 805 high-quality accurate SNP locus information (filtering condition: Dp2-miss0.5-maf0.01) can be used for subsequent population genetic diversity analysis.
|
Figure 1 The detection figures of DNA samples from Picea brachytyla Note: M-1: Trans 2k Plus DNA Marker; M-2: Trans 15k Plus DNA Marker, S: Standard, 6~20: The test samples of P. brachytyla |
1.2 Genetic phylogenetic tree analysis of natural population of Picea brachytyla
Among the 90 individuals of Picea brachytyla, the group composed of 28 individuals TPG2~TPG15, JWC1~JWC8 and JWC10~JWC15 was first separated from other groups to form group POP1 (Figure 2). In addition to 14 individuals of QZG, group POP2 also included 11 individuals of YLG, 1 individual of JWC, 1 individual of SXC and 1 individual of TPG. The individuals from the GGL sampling population were all clustered into the group POP3. In addition, it also included 14 individuals of SXC, 4 individuals of YLG and 1 individual of QZG. The genetic distances of TPG, QZG and GGL were far, and the genetic distances between TPG and JWC population, GGL and SXC population, QZG and YLG population were also close (Figure 2).
|
Figure 2 Phylogenetic tree based on SNP data of 90 P. brachytyla individuals Note: Red: The samples of Gonggangling; Blue: The samples of Jiawuchi; Green: The samples of Qizhaigou; Purple: The samples of Shenxianchi; Orange: The samples of Taipinggou; Yellow: The samples of Yalonggou; POP1, POP2, POP3: 90 individuals of P. brachytyla were divided POP1, POP2, and POP3 three populations |
1.3 Genetic structure analysis of natural population of Picea brachytyla
The genetic structure of 90 individuals in 6 populations was further analyzed to verify the genetic evolution relationship between individuals in the population. When K=2, 90 individuals were divided into two subgroups, the genetic information of GGL, QZG, SXC and YLG populations came from ancestor I (dark green), and the genetic information of the other two populations (JWC and TGP) came from ancestor II (dark blue) (Figure 3). Only JWC09 samples in JWC population were divided into dark green areas, indicating that JWC09 had a far-reaching genetic evolutionary relationship with JWC population. It can be inferred that JWC09 individual genetic information came from ancestor I. When K=3, the genetic information of 34 individuals (all individuals of GGL, 13 individuals of SXC, 4 individuals of YLG and 2 individuals of QZG) came from ancestor II (dark blue), the genetic information of 29 individuals (all individuals of TPG and 14 individuals of JWC) came from ancestor III (light blue), 86.7% of QZG, 73.3% of YLG, 6.7% of JWC and 13.3% of SXC came from ancestor I (dark green).
|
Figure 3 The population structure analysis on P. brachytyla in Jiuzhaigou Note: The color in figure means subgroup, the individuals with the same color mean that these individuals were clustered into the same subgroup; K=2~8: 90 P. brachytyla could divide 2~8 subgroups |
1.4 Principal component analysis
90 Picea brachytyla individuals were divided into 3 subgroups through principal component analysis by using SNP data (Figure 4). Subgroup 1 contained individuals from GGL, YLG, SXC and QZG, mainly GGL (100.0%) and SXC (93.3%). Subgroup 2 contained individuals from JWC and TPG groups, and only one individual in both groups was clustered into other subgroups, and the other 14 individuals were all clustered in subgroup 2. Subgroup 3 mainly included individuals of QZG, YLG, TPG, SXC and JWC, mainly individuals in QZG and YLG groups, including 86.7% and 73.3% individuals respectively. The clustering of most individuals followed the principle of geographical distance. For example, most individuals in TPG and JWC groups gather together to form a subgroup (Figure 5).
|
Figure 4 Principal component analysis based on SNP data of P. brachytyla |
|
Figure 5 The corresponding picture in map of principle component analysis |
1.5 Genetic diversity analysis of natural population of Picea brachytyla
The nucleotide diversity of six natural populations of Picea brachytyla was larger than 0.4, indicating that the nucleotide diversity of natural populations of Picea brachytyla in Jiuzhaigou County was at a high level (Figure 6). The genetic variation level of Picea brachytyla population was relatively low (Table 1), and the variation range of Pi value was 0.204 (QZG)~0.225 (TPG), with an average of 0.213. In the six populations, the PIC values were above 0.900, indicating that the tested populations had a high degree of polymorphism. Shannon-Weiner index ranges from 0.324 (QZG) to 0.336 (TPG), indicating that the distribution of individuals in each population was well balanced. The variation ranges of observed heterozygosity (Ho) and expected heterozygosity (He) were 0.232 (QZG)~0.244 (JWC) and 0.190 (YLG)~0.199 (TPG), respectively, indicating that there was no significant difference among the groups.
|
Figure 6 Analysis of nucleotide diversity among populations |
|
Table 1 Estimates of genetic variability in six populations of P. brachytyla Note: Pi: Nucleotide diversity; PIC: Polymorphism information; Shannon: Shannon-Wiener index; Ho: Observed heterozygosity; He: Expected heterozygosity |
The results of correlation analysis between genetic diversity parameters and altitude, longitude and latitude showed that Ho, He and Pi of Picea brachytyla population were negatively correlated with latitude, indicating that the genetic diversity of Picea brachytyla population tended to decrease gradually with the increase of latitude (Figure 7). Ho had a weak negative correlation with longitude, while He and Pi had a positive correlation with longitude. He and Ho had a negative correlation with altitude, but not significant. Pi had a very weak negative correlation with altitude, indicating that the genetic diversity level of Picea brachytyla population in relatively high altitude areas was low.
|
Figure 7 Correlation analysis between genetic diversity of P. brachytyla populations between longitude, latitude and altitude Note: The curve in the figure mean the fitting trend of genetic index |
1.6 Population genetic differentiation
The Fst values of the six Picea brachytyla populations ranged from 0.001 (between QZG and YLG) to 0.041 (between TPG and YLG) (Table 2). Among the populations with close geographical distance, the Fst value among QZG, YLG and SXC was the lowest, and the variation range was 0.001~0.004, indicating that the degree of genetic differentiation between them was very low. The populations that were geographically distant from each other, such as JWC and QZG, had a relatively high degree of genetic differentiation. Overall, the Fst among the six populations of Picea brachytyla was at a low level, in the range of 0~0.05, indicating that the genetic differentiation among the six populations was very small and can be ignored.
|
Table 2 Pairwise comparison of genetic differentiation (Fst) among six P. brachytyla populations |
2 Discussion
Single nucleotide polymorphism (SNP) information widely exists in the genome of species, which has the characteristics of high density, genetic stability and easy automation of analysis. It is widely used in the study of genetic diversity among plant populations and individuals (Wang et al., 2019). In this study, 26 805 high-quality and high-precision SNP loci of Picea brachytyla were obtained by simplified genome sequencing technology, and these SNP loci were used to study the genetic diversity of 6 natural populations of Picea brachytyla. The population stratification can be statistically corrected by estimating the number of genetic ancestors based on multipoint genotype data (Alexander et al., 2009). Population genetic structure analysis clusters according to the differences between individuals. Although clustering will reduce the genetic diversity information in each population, it also makes individuals tend to the degree of their clustered population, which is helpful to comprehensively understand their genetic diversity and genetic structure information from the population and individual levels (Wang et al., 2012). Genetic genotype analysis and genetic structure analysis study population genetic diversity from different sides, but the two analysis methods can verify and supplement each other to ensure the stability and reliability of the analysis results. The level of genetic diversity of woody plants with a wide distribution area for many years is generally at a high level (Zhu et al., 2013; Shen et al., 2014). In this study, the average Ho of six natural populations of Picea brachytyla was 0.235 and the average He was 0.194, which showed the low level of genetic diversity of Picea brachytyla population in Jiuzhaigou. It was speculated that the reason may be that the distribution altitude of Picea brachytyla is high and the climate change is complex, resulting in insufficient gene exchange among populations; Secondly, habitat fragmentation reduces gene exchange among populations of Picea brachytyla, and self breeding reduces genetic diversity. The comprehensive analysis showed that the genetic diversity of the natural population of Picea brachytyla showed a downward trend from east to west with the increase of altitude, and an upward trend from south to north with the decrease of altitude. This conclusion was different from the research results of other tree species. For example, the genetic diversity of natural population of Sophora moorcroftiana increases with the increase of altitude (Liu, 2019). The results of Wang et al. (2019) on the genetic diversity of Litsea populifolia population showed that altitude had little effect on the genetic diversity of Litsea populifolia. This may be related to the biological characteristics of different tree species: the higher altitude areas show strong natural selection pressure due to the relatively low temperature, which affects the gene exchange between populations, resulting in the relatively low genetic diversity of Picea brachytyla population in the higher altitude areas. Based on the analysis of genetic diversity and the investigation of population habitat, the results were as follows: (1) the genetic diversity of low altitude population of Picea brachytyla was relatively high; (2) Habitat fragmentation caused by human activities such as over exploitation, grazing and road construction may also reduce the genetic diversity of Picea brachytyla. Seed banks have a direct impact on population dynamics (Harper, 1977). In the field investigation, few seeds and seedlings were found in the natural population of Picea brachytyla, indicating that the natural regeneration ability of Picea brachytyla is very poor, which may be one of the main reasons for the low level of genetic diversity of Picea brachytyla natural population. The ability of plant population to resist the threat of adverse environment is directly related to the level of genetic diversity of the population (Spielman et al., 2004). Therefore, we should strengthen the protection of Picea brachytyla natural population in Jiuzhaigou County to avoid its decline.
The asynchronous flowering period between individuals, the difference in the amount of male and female flowers or other changes in environmental factors will limit the pollen transmission of cross pollinated plants, break the random mating system within the population, and significantly increase the inbreeding rate (Murawski and Hamrick, 1991; Yang et al., 2016; Yang et al., 2017). In this study, each population still has enough heterozygotes (Ho>He). Population nucleotide diversity analysis and genetic differentiation analysis also showed that Picea brachytyla in Jiuzhaigou has high intra population genetic variation, but there is relatively little gene exchange between populations. The same genus Picea likiangnsis also has a low level of genetic diversity (He=0.250) (Peng and Wu, 2011). Picea schrenkiana var. tianschanica has high genetic diversity, and its Ho and He were 0.4923 and 0.5907 respectively (Ji, 2012). It can be seen that Picea brachytyla showed low genetic diversity and low population genetic differentiation in Picea. This is related to the life history of the tree species and the unique geographical factors of Jiuzhaigou. The natural population of Picea brachytyla in Jiuzhaigou County is basically distributed continuously, and the gene exchange between populations is relatively frequent or unrestricted, which makes the low degree of genetic differentiation among the six natural populations of Picea brachytyla in this study. Although most natural populations of Picea brachytyla are distributed continuously, the number of individuals in most populations is small, and most of them are adult trees, which may be the main factor leading to the low level of genetic diversity of Picea brachytyla in Jiuzhaigou.
3 Materials and Methods
3.1 Materials
This study was conducted in Jiuzhaigou County, Sichuan Province in October 2018. According to the preliminary investigation results of Picea brachytyla, six natural populations were selected for population sampling in the distribution area, and 15 plants were selected for sampling in each population, with an interval of about 50 m between adjacent plants (Table 3). Collect the young needles of Picea brachytyla, dry them with silica gel and store them in - 80℃ ultra-low temperature refrigerator for later use.
|
Table 3 New ICT based fertility management model in private dairy farm India as well as abroad |
3.2 Extraction of genomic DNA
Plant genomic DNA Extraction Kit (TaKaRa, Dalian) was used to extract genomic DNA of Picea brachytyla. 1% agarose gel electrophoresis, Nanodrop and Qubit were used to detect the integrity, purity and concentration of DNA of Picea brachytyla, so as to ensure that the extracted DNA could meet the requirements of building library.
3.3 GBS library construction and sequencing
Firstly, the genome was digested by restriction enzyme to obtain the appropriate Marker density. The digested fragments were added with P1 and P2 Adapter with barcode, which can complement the digested DNA gap, and then all 90 individual samples were amplified by PCR. Finally, the PCR products were electrophoretically recovered and the required fragments were used to construct the library. After the library was successfully constructed, the constructed library was sequenced by Illumina Hiseq platform.
3.4 SNP detection
After obtaining sequencing data, SAMTOOLS v1.4 software was used to detect single nucleotide polymorphism (SNP) information sites on the sequencing data of 90 individual samples of Picea brachytyla (Li et al, 2009). SAMTOOLS software was used to detect the common SNP sites. After filtering, high-quality SNP sites were finally obtained for subsequent analysis.
3.5 Genetic structure analysis
Based on the SNP locus information of 90 individuals in Picea brachytyla population, the genetic distance between individuals was estimated by TreeBest software (Bootstrap=1 000), and the phylogenetic tree of 90 individuals based on genetic structure analysis was constructed by Neighbor-joining method, GCTA software (http://cnsgenomics.com/software/gcta/pca.html) was used to calculate the eigenvector and eigenvalue, and R language was used to draw the PCA distribution map. The PLINK input file was constructed according to the SNP locus information of 90 individuals. The population genetic structure information of natural population of Picea brachytyla was constructed by frappe software, and its population genetic structure was analyzed.
3.6 Genetic diversity analysis method
The observed heterozygosity (Ho) and expected heterozygosity (He) of Picea brachytyla natural population were calculated by Arlequin software. AMOVA 1.55 software and DnaSP v5 software were used to analyze the molecular variance and nucleotide diversity (Π) of the natural population of Picea brachytyla. The calculation formula of Ho was: number of observed heterozygotes/total number of samples, that is, Ho = number of observed heterozygotes/90. He was calculated according to the formula provided by Nei (1978):
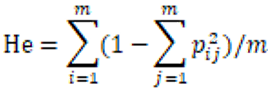
Where, m is the number of loci and pij is the frequency of allele j at locus i.
Authors’ contributions
GYJ and MJY were the experimental designers and executors of this study; PJ and LXQ completed data analysis and wrote the first draft of the manuscript; LXQ participated in the experimental design and analyzed the experimental results; MJY and GYJ were the conceivers and principals of the project, guiding the experimental design, data analysis, manuscript writing and modification. All authors read and approved the final manuscript.
Acknowledgments
This study was funded by “Demonstration of Genetic Diversity Protection of Picea brachytyla Germplasm Resources” (201827).
Alexander D.H., Novembre J., and Lange K., 2009, Fast model-based estimation of ancestry in unrelated individuals, Genome Res., 19(9): 1655-1664
https://doi.org/10.1101/gr.094052.109
PMid:19648217 PMCid:PMC2752134
Harper J.L., 1977, The population biology of plants, Academic Press, London, UK, pp.35-37
Ji X.B., 2012, Genetic diversity of Picea schrenkiana var. tianschanica in different altitude by EST-SSR markers analysis, Thesis for M.S., Agricultural University of Hebei, Supervisors: Wang J.M., and Yang M.S., pp.10-30
Liu Y., 2019, Population genetic characteristics and representative populations’ phenotypic traits of Sophora moorcroftiana, Thesis for M.S., Northwest A&F University, Supervisors: Wang C.Y. pp.20-45
Lu W.H., Xiong T., Wang J.Z., Zhang L., Qi J., Luo J.X., and Xie Y.J., 2018, Genetic diversity of 1st generation breeding population in Eucalyptus urophylla, Jiyinzuxue yu Yingyong Shengwuxue (Genomics and Applied Biology), 37(6): 2505-2517
Lyu Z.H., and Guo H.C., 2018, Comparison of genetic diversity analysis by RSAP, SSR, and SRAP markers in potato, Jiyinzuxue yu Yingyong Shengwuxue (Genomics and Applied Biology), 37(6): 2544-2550
Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., and Durbin R., 2009, The sequence alignment/map format and SAMtools, Bioinformatics, 25(16): 2078-2079
https://doi.org/10.1093/bioinformatics/btp352
PMid:19505943 PMCid:PMC2723002
Murawski D.A., and Hamrick J.L., 1991, The effect of the density of flowering individuals on the mating systems of nine tropical tree species, Heredity, 67(2): 184
https://doi.org/10.1038/hdy.1991.76
Nei M., 1978, Estimation of average heterozygosity and genetic distance from a small number of individuals, Genetics, 89(3): 583-590
https://doi.org/10.1093/genetics/89.3.583
PMid:17248844 PMCid:PMC1213855
Poland J.A., Brown P.J., Sorrells M.E., and Jannink J.L., 2012, Development of high-density genetic maps for barely and wheat using a novel two-enzyme genotyping-by-sequencing approach, PLoS One, 7(2): e32253
https://doi.org/10.1371/journal.pone.0032253
PMid:22389690 PMCid:PMC3289635
Peng X.L., and Wu W.Z., 2011, Genetic variation and phytogeography of Picea likiangensis inferred from RAPD markers, Zhiwu Yanjiu (Bulletin of Botanical Research), 31(4): 436-442
Shen D.F., Bo W.H., Xu F., and Wu R.L., 2014, Genetic diversity and population structure of the Tibetan poplar (Populus szechuanica var. tibetica) along an altitude gradient, BMC Genet., 15(S1): 11
https://doi.org/10.1186/1471-2156-15-S1-S11
PMid:25079034 PMCid:PMC4118629
Spielman D., Brook B.W., and Frankham R., 2004, Most species are not driven to extinction before genetic factors impact them, Proc. Natl. Acad. Sci. USA, 101(42): 15261-15264
https://doi.org/10.1073/pnas.0403809101
PMid:15477597 PMCid:PMC524053
Taranto F., Agostino N.D., Greco B., Cardi T., and Tripodi P., 2016, Genome-wide SNP discovery and population structure analysis in pepper (Capsicum annuum) using genotyping by sequencing, BMC Genomics, 17(1): 943
https://doi.org/10.1186/s12864-016-3297-7
PMid:27871227 PMCid:PMC5117568
Wang X., Gao M., Wu L.W., Wang Y.D., Liu H.C., and Chen Y.C., 2019, A study on population genetic diversity among Litsea populifolia (Hemsl.) gamblein mount Emei area, Zhiwu Yichuan Ziyuan Xuebao (Journal of Plant Genetic Resources), 20(2): 359-369
Wang H.P., Simon P.W., Li X.X., Cheng J.Q., Shen D., Song J.P., Qiu Y., and Zhang X.H., 2012, Diversity and genetic structure of garlic (Allium sativum L.) germplasm resource in China, Zhongguo Nongye Kexue (Scientia Agricultura Sinica), 45(16): 3318-3329
Yang H.B., Zhang R., and Zhou Z.C., 2016, Genetic diversity and mating system in a seed orchard of Schima superba, Linye Kexue (Scientia Silvae Sinicae), 51(12): 66-73
Yang H.B., Zhang R., Song P., and Zhou Z.C., 2017, Flowering phenology and synchronization of clones among plant ages in a seed orchard of Schima superba, Linye Kexue Yanjiu (Forest Research), 30(4): 551-558
Zhu J.H., Pan L.M., Qin X.Q., Peng H.X., Wang Y., and Han Z.H., 2013, Analysis on genetic relations in different ecotypes of longan (Dimocarpus longan Lour.) germplasm resources by ISSR markers, Zhiwu Yichuan Ziyuan Xuebao (Journal of Plant Genetic Resources), 14(1): 65-69
. PDF(1270KB)
. HTML
Associated material
. Readers' comments
Other articles by authors
. Jingyan Mo
. Jian Peng
. Yunjie Gu
. Xiaoqing Li
Related articles
. Picea brachytyla
. GBS
. Genetic diversity
. Genetic conservation
Tools
. Email to a friend
. Post a comment
.png)
.png)
.png)
.png)
.png)
.png)
.png)
.png)
.png)
.png)


